HiDome
The core objective of this delivery is to allow researchers to create patient cohorts by setting constraints on components in high dimensional data sets, e.g. select all patients with expression value of a particular gene greater than a user-selected threshold. Additionally, the values for these components can be viewed like regular numerical concepts in the Summary Statistics and Grid View tabs. To this end, the tranSMART web application will be extended with GUI features that will allow researchers to specify constraints over these components. By allowing researchers to directly compare clinical data with high dimensional data, the integration of HiDome in tranSMART will expedite translational research.
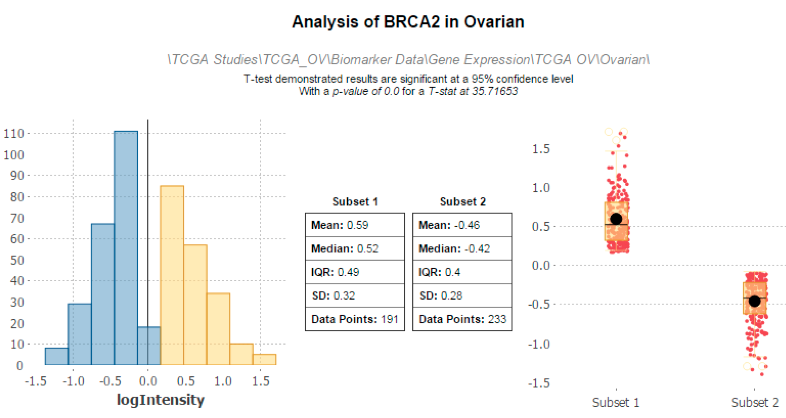
HiDome can be used in a variety of ways. On a HiDome-enabled installation of Transmart you can drag and drop a high-dimensional data node either into the cohort selection tool, in the summary statistics tabs or the grid view tab to access the high-dimensional data. In cohort selection, it can be used to further refine patient cohorts. For instance, you may be interested to divide your patients into a group with low expression of the TNF gene, and a group with high expression of the TNF gene. You can combine these constraints with clinical data constraints such as age. You can also use HiDome to explore the distribution of expression values in patient subsets. To do this, simply drag the high dimensional node to the summary statistics tab after creating your subsets, and indicate the gene of interest. If your study contains multiple high dimensional samples, multiple platforms, or even multiple data types, you can use HiDome to explore relations between all of these. For example, using HiDome you can create two subsets based on gene expression, and explore the effect this has the distribution of metabolomics or proteomics data.
Contact Us!
Please feel free to contact Denny Verbeeck (dverbeec@its.jnj.com) with any bug reports, feature requests or other comments relating to HiDome.Acknowledgements
The authors would like to acknowledge the contributions of the IMI and EFPIA. Special thanks to Johnson & Johnson, Bayer, Imperial College London, University of Luxembourg and TheHyve for their respective support on HiDome.Disclaimer
HiDome is offered to the public as freely available resource, for non-commercial research use. Some aspects of this experimental module may still be under development, and there are no warranties about the completeness, reliability, accuracy, and security of the software package. Please bear this in mind, especially if you wish to analyse personal and/or confidential data.
